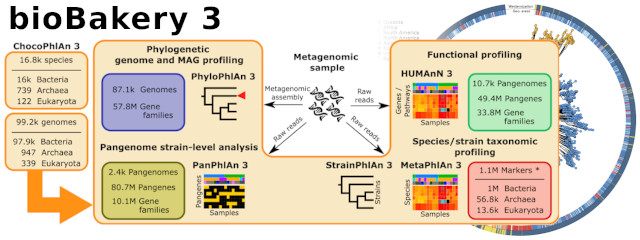
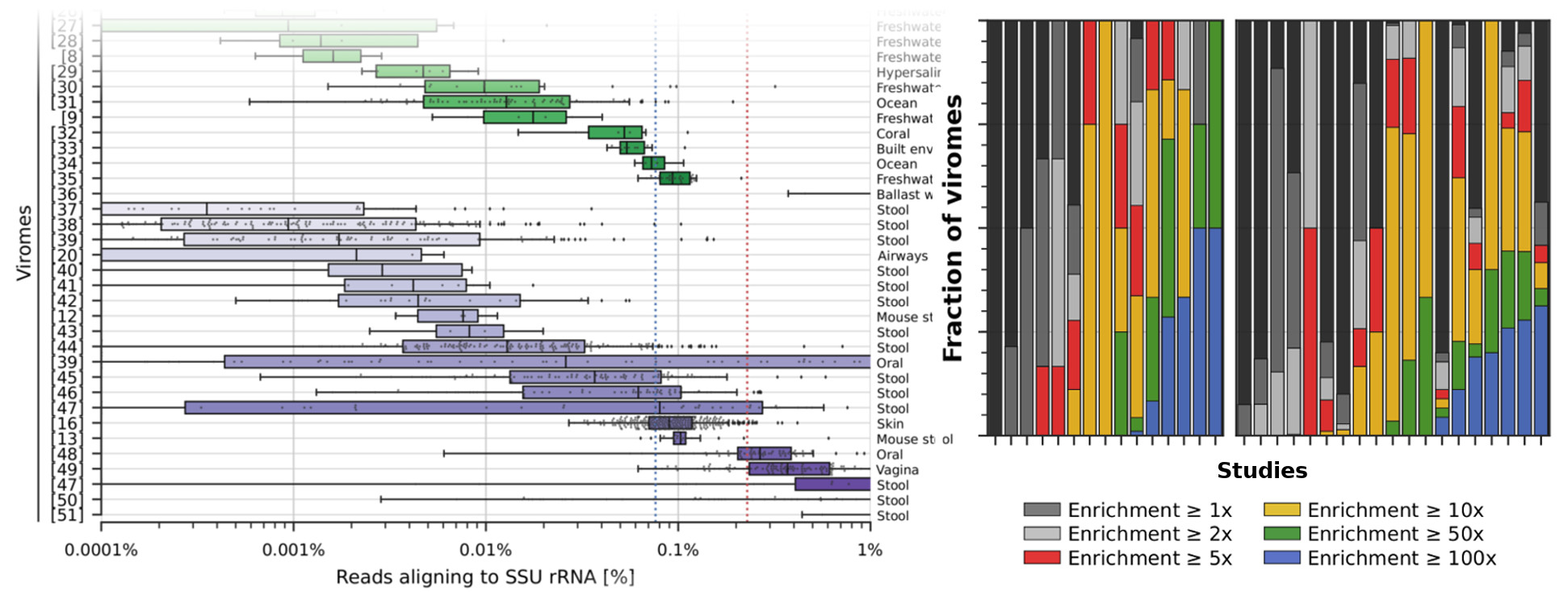
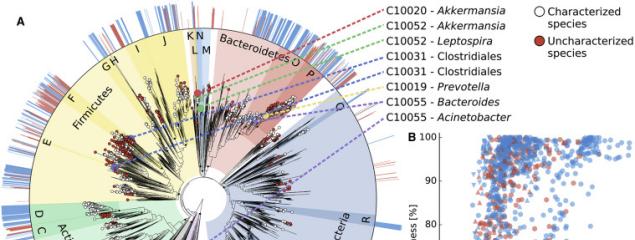
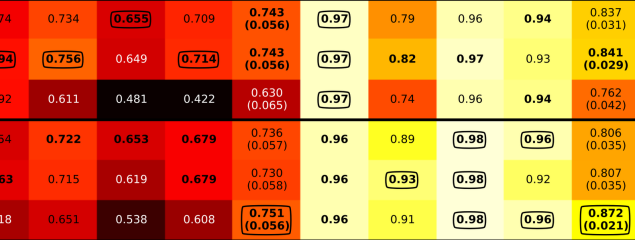
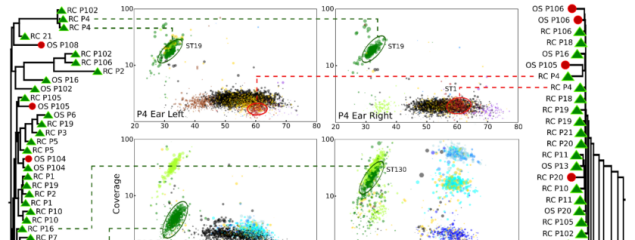
Release of a new version of MetaPhlAn –MetaPhlAn v4.2.2– incorporates taxonomic profiling of long-read metagenomes for the first time and includes a new version of the MetaPhlAn db (vJan25_202503) containing >21k new SGBs.
Meta'omics for hacking the human microbiome
Our body harbours ~100 trillion microbes which outnumber our own cells. This microbial diversity (the "microbiome") and its functions are still largely uncharachterized but we can now try to mine it using cultivation-free sequencing-based metagenomic tools.
We employ experimental meta'omic tools and novel computational approaches to study the diversity of the microbiome and its role in human dysbiosis and infections. Our projects bring together computer scientists, microbiologists, statisticians, and clinicians.
Main research directions
Next generation computational metagenomic tools. The potential of metagenomics is only partially expressed due to computational challenges. We are working on novel methods to profile microbiomes at increased resolution (e.g. strains) and perform large-scale comparative genomics on uncharacterized microbes.
Integrative and machine learning meta'omic approaches. We develop new machine learning tools to cope with the variability and dimensionality of microbiome profiles and provide clinically relevant signatures by integrating complementary meta'omic approaches (e.g. metatranscriptomics or metaproteomics).
Microbiome-pathogen interaction in human infections. The role of the microbiome in the acquisition and development of infections is largely unknown. By coupling longitudinal pathogen/microbiome sequencing we aim understand how the microbiome can modulate the virulence profile and antibiotic resistance of human infections.
Microbiome transmission. We study how microbes can be transmitted between different environments with specific focus on how members of the microbiome are vertically transmitted from mothers to infants at birth and horizontally between family members or individuals sharing the same environment. .
Our Keywords (from our papers)