GraPhlAn
GraPhlAn is a software tool for producing high-quality circular representations of taxonomic and phylogenetic trees. It focuses on concise, integrative, informative, and publication-ready representations of phylogenetically- and taxonomically-driven investigation.
Software repository and supporting material
The software repository of GraPhlAn is at: https://github.com/biobakery/graphlan
The supporting user group is at: https://forum.biobakery.org/c/Infrastructure-and-utilities/GraPhlAn/17
A first comprehensive tutotial is at: https://github.com/biobakery/graphlan/wiki/Home
A tutorial in Biobakery is at: https://github.com/biobakery/biobakery/wiki/graphlan
A Galaxy interface is at: http://huttenhower.sph.harvard.edu/galaxy/root?tool_id=graphlan
For comments and question please refer to the user group linked above or contact us
Citation
Compact graphical representation of phylogenetic data and metadata with GraPhlAn
PeerJ, 2015 10.7717/peerj.1029
1 Centre for Integrative Biology (CIBIO), University of Trento, Italy
2 Biostatistics Department, Harvard School of Public Health, USA
2 Broad Institute of MIT and Harvard, USA
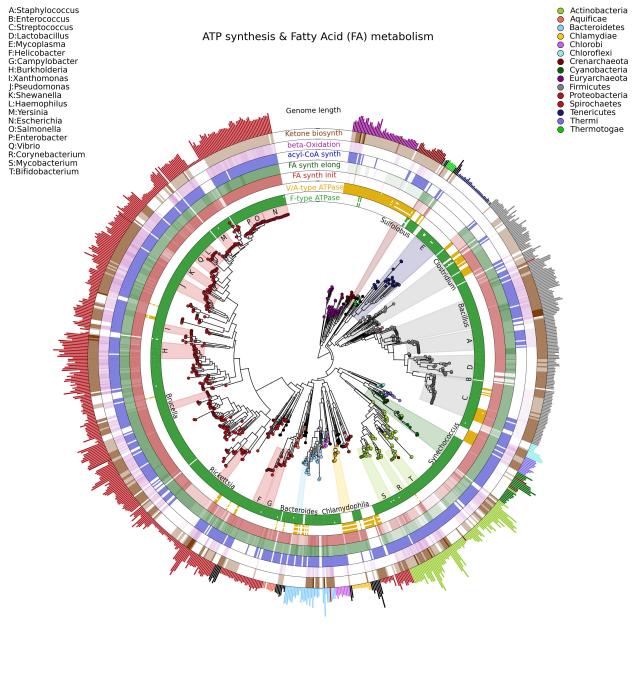
Example
An example figure is below (more on the example folder in the repository)