Software repository and supporting material
Software repository of PanPhlAn:
https://github.com/segatalab/panphlan
Available species pangenome databases:
https://github.com/segatalab/panphlan/wiki/Pangenome%20databases
The PanPhlAn tutorial:
https://github.com/segatalab/panphlan/wiki
User support (email-based group and discussion forum):
https://forum.biobakery.org/c/Microbial-community-profiling/PanPhlAn
For comments and question please write to our
user support group
or contact directly the Segata
lab.
Publication
If you find this tool useful in your research, please cite our
papers:
Francesco Beghini1
Lauren J McIver2
Aitor Blanco-Mìguez1
Leonard Dubois1
Francesco Asnicar1
Sagun Maharjan2,3
Ana Mailyan2,3
Andrew Maltez Thomas1
Paolo Manghi1
Matthias Scholz2,3
Mireia Valles-Colomer1
George Weingart2,3
Yancong Zhang2,3
Moreno Zolfo1
Curtis Huttenhower2,3
Eric A Franzosa2,3
Nicola Segata1,5
Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3
eLife(2021)
https://doi.org/10.7554/eLife.65088
1 Department CIBIO, University of Trento, Italy
2 Harvard T. H. Chan School of Public Health, Boston, MA, USA
3 The Broad Institute of MIT and Harvard, Cambridge, MA, USA
4 Department of Food Quality and Nutrition, Research and Innovation Center, Edmund Mach Foundation, Italy
5 IEO, European Institute of Oncology IRCCS, Milan, Italy
Matthias Scholz1,*
Doyle V. Ward2,*
Edoardo Pasolli1,*
Thomas Tolio1
Moreno Zolfo1
Francesco Asnicar1
Duy Tin Truong1
Adrian Tett1
Ardythe L. Morrow3
Nicola Segata1
Strain-level microbial epidemiology and population genomics from shotgun metagenomics
Nature Methods 13, 435–438, 2016.
10.1038/nmeth.3802
* Equal contribution
1 Centre for Integrative Biology, University of Trento, Trento, Italy
2 Center for Microbiome Research, University of Massachusetts Medical School, Worcester, Massachusetts, USA
3 Department of Pediatrics, Perinatal Institute, Cincinnati, Ohio, USA
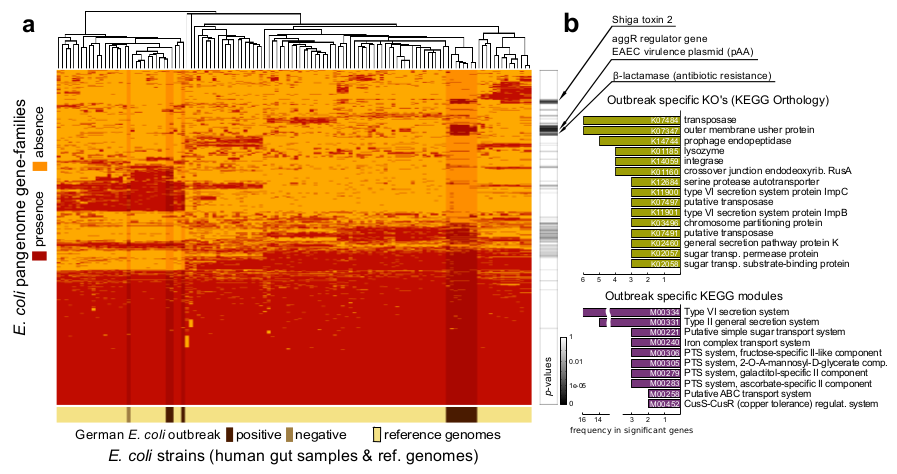
Example of E. coli strain profiling
Characterization of the German 2011 E. coli outbreak strain
PanPhlAn profiling
of the German outbreak metagenomes using a reference database in
which the target outbreak genome is missing. (a) Hierarchical
clustering. The heatmap displays presence/absence gene-family
profiles of 110 reference strains (bright colored columns) and of 12
metagenomically detected strains (darker columns). Most outbreak
samples cluster together due to almost identical profiles (right),
with four samples (left) showing different profiles due to the
presence of additional dominant E. coli strains overlying
the target outbreak strain. (b) Functional analysis of
outbreak-specific gene-families (Fisher exact test) confirmed that
the outbreak strain is a combination of a EAEC pathogen
(pAA plasmid) with acquired Shiga toxin and antibiotic resistance
genes, complemented with a set of enriched virulence-related
functions and pathway modules.