StrainPhlAn - A metagenomic strain-level population genomics tool
StrainPhlAn is a computational tool for performing metagenomic strain-level population genomics on large metagenomic datasets by profiling microbes from known species with strain level resolution and providing comparative and phylogenetic analyses of strains retrieved from metagenomic samples.
StrainPhlAn takes as input a set of metagenomic samples (in fastq format), reference genomes (in fasta) and then reconstructs the sample-specific strains of all species present in the samples and the genome-specific strains from reference genomes. The figure below shows the StrainPhlAn framework and its application for Prevotella copri on a subset of the samples considered from nine studies.
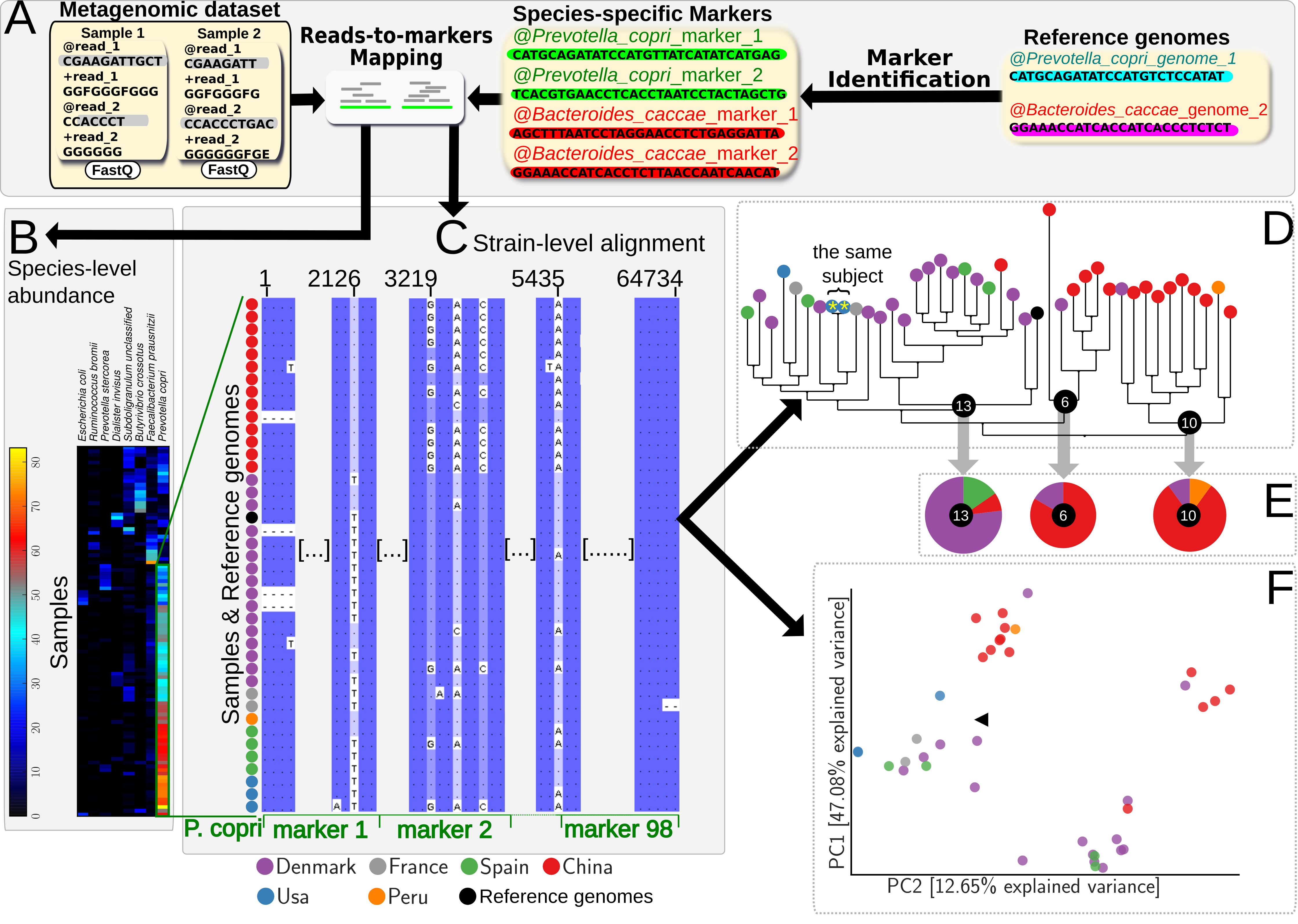
StrainPhlAn seful links
- The StrainPhlan GitHub repository
- The StrainPhlan 4.1 documentation/tutorial
- The StrainPhlan 4.0 tutorial
- The StrainPhlan user support
Citation
If you use StrainPhlAn, please cite the following: papers:
bioRxiv(2022) https://doi.org/10.1101/2022.08.22.504593
1 Department CIBIO, University of Trento, Italy
2 Harvard T. H. Chan School of Public Health, Boston, MA, USA
3 The Broad Institute of MIT and Harvard, Cambridge, MA, USA
4 Department of Experimental Oncology, IEO European Institute of Oncology IRCCS, Milan, Italy
5 Centre for Microbiology and Environmental Systems Science, University of Vienna, Austria
6 Zoe Global, London, UK
7 Department of Nutritional Sciences, King’s College London, London, UK
8 Department of Twin Research, King’s College London, London, UK
9 Department of Agricultural Sciences, University of Naples, Naples, Italy
Microbial strain-level population structure and genetic diversity from metagenomes
Genome Research(2017) 10.1101/gr.216242.116
1 Centre for Integrative Biology, University of Trento, Trento, Italy
2 Harvard T.H. Chan School of Public Health


